After successful implementation by our DBPs, the Community Engagement arm of the Center begins the process of making the technology sustainably available to the broader community through non-Center entities such as repositories or companies. While this sustainable distribution is being put into place, direct distribution in a limited fashion may be provided by the Center to some "early adopters" who ask us for access to the technology as soon as it has been proven reliable.
Technology Optimization Projects
Each TOP is focused on optimizing a subset of GCE tools that can have great impact in advancing biomedical research, but need to be made more robust and efficient in terms of the incorporation of certain types of non-canonical amino acids (ncAAs) and in terms of protocols for optimally handling and using the GCE-produced designer proteins.
The advances in Genetic Code Expansion technology targeted by GCE4All are organized into two TOPs:
- TOP-1 focuses on ncAAs for bioorthogonal ligations
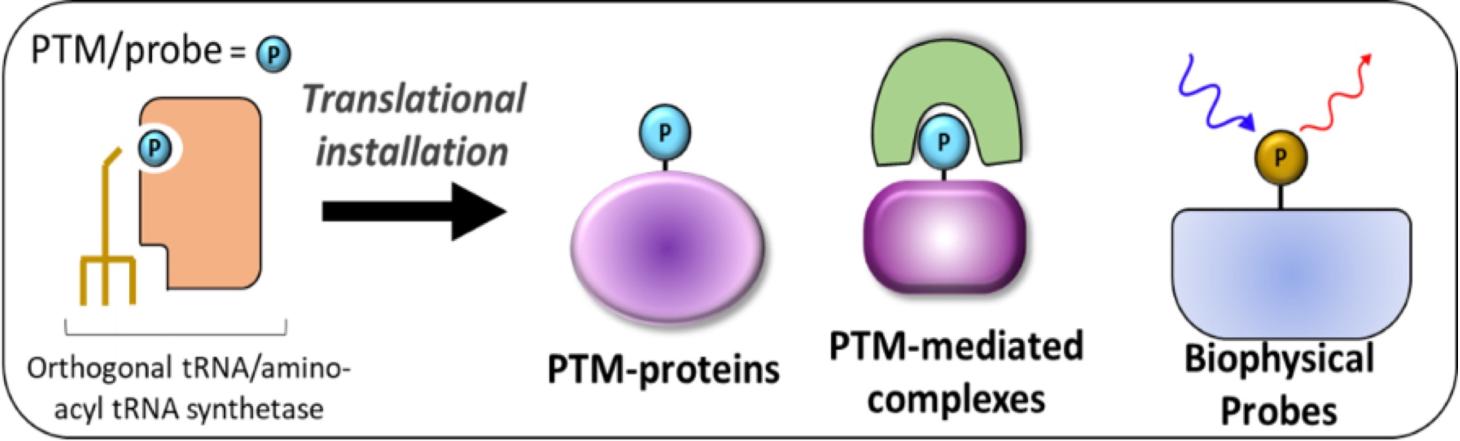
- TOP-2 focuses on ncAAs that can serve as biochemical probes as well as ncAAs that represent post-translational modifications
As the TOPs develop technologies, GCE4All investigators will gain valuable experience regarding how to most effectively develop, optimize and implement GCE tools for any given ncAA. As a synergistic outcome, the combined experience of the TDPs will be leveraged to develop robust approaches and protocols for GCE tool development. We call these “GCE bridges,” and our vision is that these bridges will give any molecular biology lab the ability to develop – from scratch – a full set of efficient GCE tools for a novel ncAA of interest. The four GCE bridges to be built are:
- The bridge to access a new ncAA through developing the needed RS/tRNA pair
- Given an RS/tRNA pair, the bridge to optimize ncAA-protein expression in E. coli
- Given an RS/tRNA pair, the bridge to optimize ncAA-protein expression in mammalian cells
- Given an RS/tRNA pair with optimized expression, the bridge to create a stable GCE mammalian cell line
TOP-1: GCE Technologies for Bioorthogonal ligations
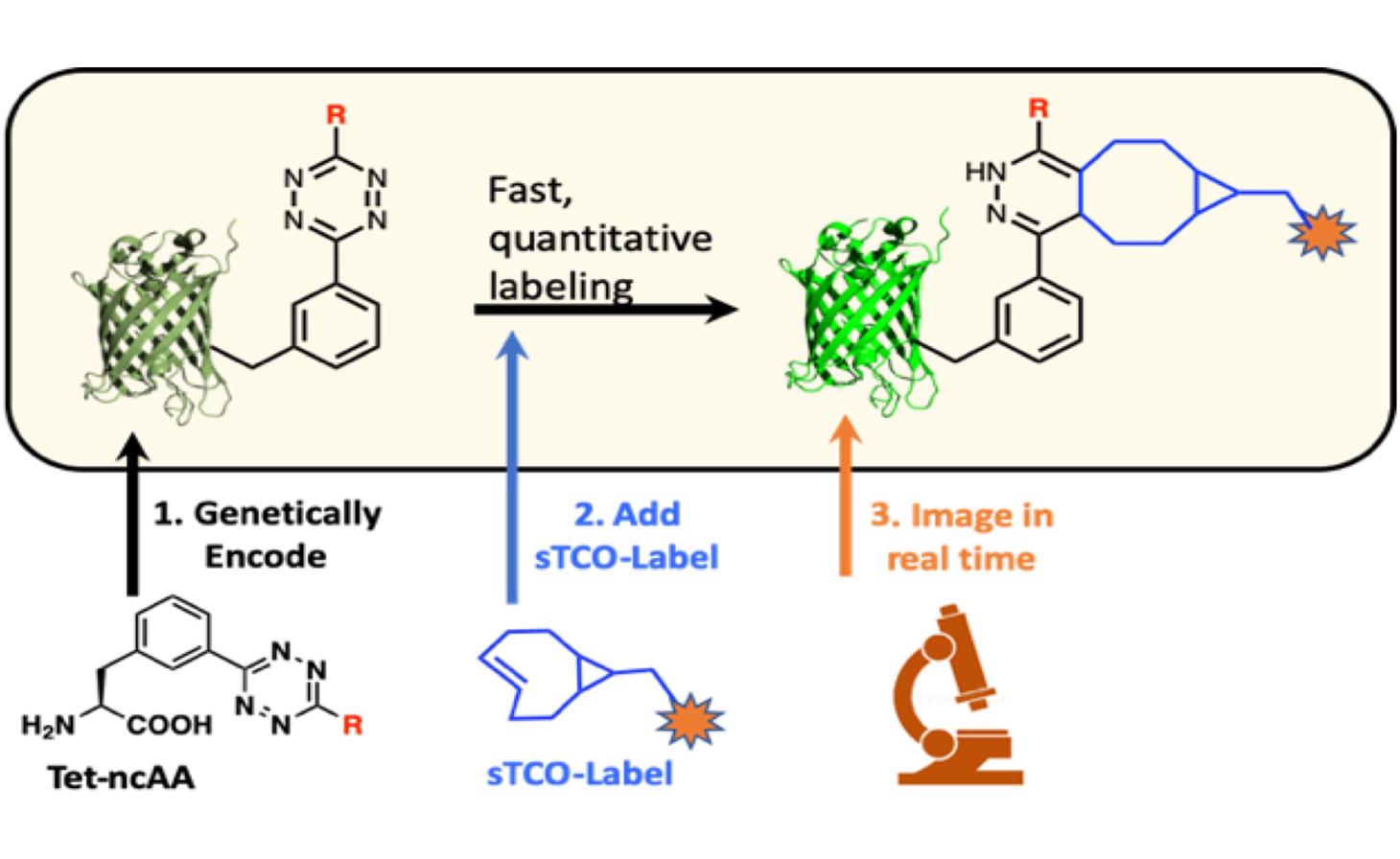
Current TOP-1 projects focus on incorporating tetrazine amino acids into proteins and reacting them with trans-cyclooctane (TCO) reactive groups that can be attached to a fluorescent dye or via a linker to another protein or surface (building on the chemistry reported by Blizzard et al 2015). GCE technologies enabling efficient tetrazine amino acid incorporation into proteins in both E. coli and mammalian expression hosts will be optimized alongside conditions for high-yield reactions with TCO-activated ligation targets. When expressed in mammalian cells, Tet-proteins react intracellularly providing unprecedented abilities to modify and study proteins in their physiological settings. Once optimized, GCE bioorthogonal ligations will enable attachment of larger fluorophores further enabling the addition of polymers, drugs, imaging agents, or other biomolecules and materials to ultimately impact a wide range of science.